Overall Value
METAGENE-1 bridges the gap between public health surveillance and advanced genomic AI. Its focus on real, messy, and diverse metagenomic data, rather than pristine lab sequences, makes it an unmatched tool for understanding the true biological signals in our environment. Whether you’re monitoring a potential outbreak or studying microbial ecosystems, METAGENE-1 brings precision, scalability, and early-warning capabilities to your research workflow.
Features
• Analyze noisy, short metagenomic reads from complex samples
• Detect potential pathogen anomalies in genomic data streams
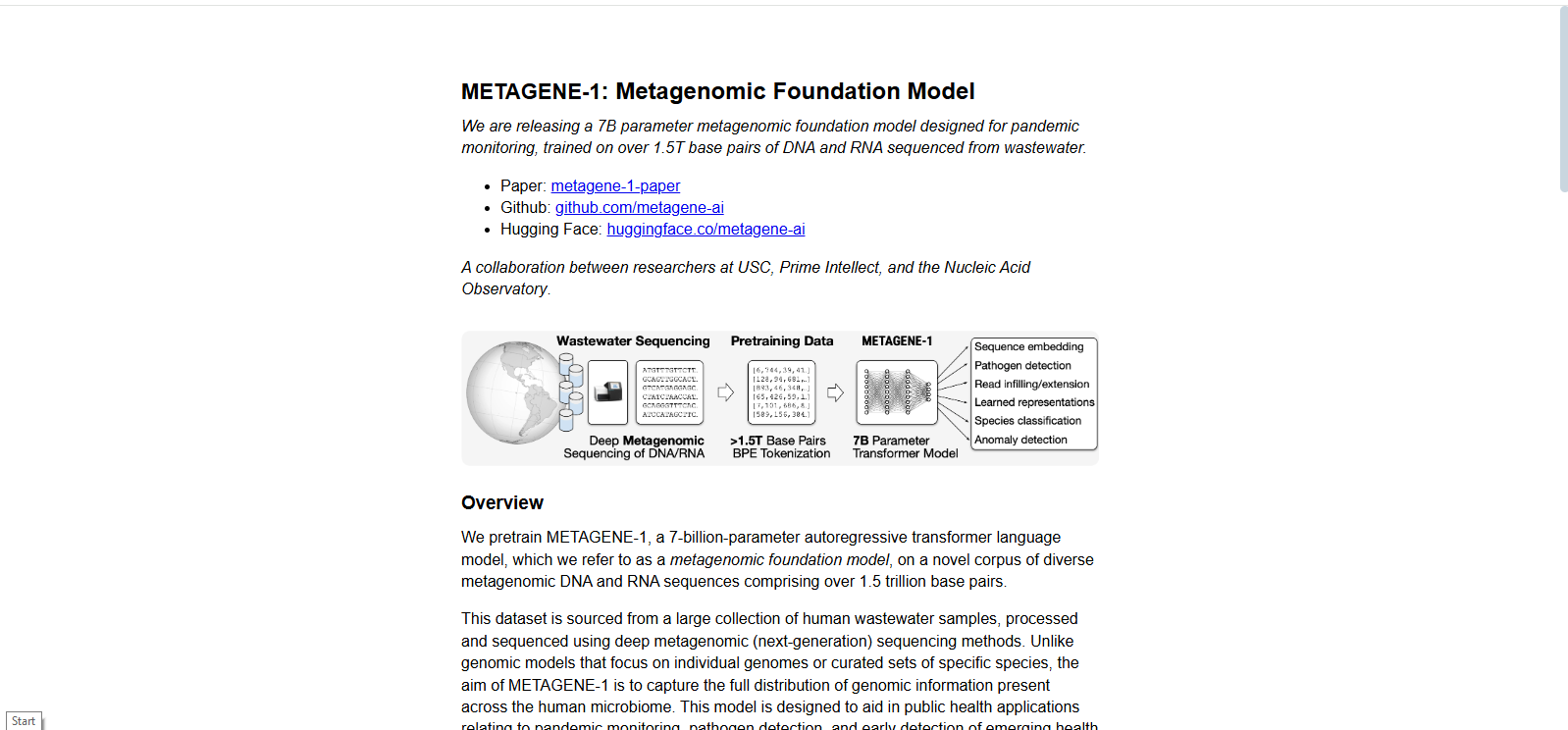
• Pretrained on 1.5T+ base pairs from human wastewater metagenomes
• Custom BPE tokenizer optimized for microbial DNA/RNA patterns
• Embeds diverse genomic fragments for unsupervised representation learning
• 512-token context window fine-tuned for high-speed scanning tasks
• Foundation model architecture built for generalization across species
• Emphasizes safety and misuse resistance by design
• Compatible with standard bioinformatics pipelines and benchmarking datasets
Use Cases
✔️ Flag early signals of pandemics through environmental surveillance
✔️ Model microbial population shifts in public wastewater
✔️ Detect rare or novel pathogens without predefined species templates
✔️ Train downstream diagnostic tools for precision health monitoring
✔️ Study metagenomic patterns across urban or geographic populations
✔️ Assist national biosurveillance systems and academic biolabs
Technical Specifications
- 7B parameter transformer model
- Autoregressive training on uncurated metagenomic sequences
- Trained on 1.5T+ base pairs of DNA/RNA from wastewater samples
- Custom byte-pair encoding (BPE) tokenizer tailored for microbial data
- Optimized for reads in the 100–300 base pair range
- 512-token context window for short-sequence inference
- Released as open-source by USC, Prime Intellect, and the Nucleic Acid Observatory
- Benchmarked on pathogen detection and genomic embedding tasks
 Stay ahead of the curve with METAGENE-1’s open-source metagenomic intelligence model Detect pathogens, analyze anomalies, and model short genomic reads like never before
Stay ahead of the curve with METAGENE-1’s open-source metagenomic intelligence model Detect pathogens, analyze anomalies, and model short genomic reads like never before
FAQs
Not directly. It’s optimized for population-scale surveillance, not individual diagnostics. However, it can support upstream signals that feed into clinical pipelines.
Most models are trained on curated genomes from specific species. METAGENE-1 learns from messy, real-world metagenomes, giving it broader generalization and anomaly detection capabilities.
No. METAGENE-1 is intentionally limited in sequence generation scope. Its 512-token context and safety-first design reduce the potential for misuse in synthetic applications.
Epidemiologists, public health researchers, microbiome scientists, and anyone working in bio-surveillance, environmental monitoring, or genomic data science.
Standard model deployment tools like Hugging Face Transformers are sufficient. For high-throughput tasks, GPU-accelerated inference is recommended.
Conclusion
METAGENE-1 redefines how we understand and monitor microbial life at scale. With unmatched exposure to real-world genomic diversity, it equips researchers and public health officials to spot biological anomalies before they escalate. As pandemics and environmental threats loom, METAGENE-1 is more than a model—it’s a biosurveillance engine for the genomic age.



 Stay ahead of the curve with METAGENE-1’s open-source metagenomic intelligence model Detect pathogens, analyze anomalies, and model short genomic reads like never before
Stay ahead of the curve with METAGENE-1’s open-source metagenomic intelligence model Detect pathogens, analyze anomalies, and model short genomic reads like never before